Note
Go to the end to download the full example code
Diagonal Gauss-Newton Second order optimizer
A simple second-order optimizer with BackPACK on the classic MNIST example from PyTorch. The optimizer we implement uses uses the diagonal of the GGN/Fisher matrix as a preconditioner, with a constant damping parameter;
where
Let’s get the imports, configuration and some helper functions out of the way first.
import matplotlib.pyplot as plt
import torch
from backpack import backpack, extend
from backpack.extensions import DiagGGNMC
from backpack.utils.examples import get_mnist_dataloader
BATCH_SIZE = 128
STEP_SIZE = 0.05
DAMPING = 1.0
MAX_ITER = 200
PRINT_EVERY = 50
DEVICE = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
torch.manual_seed(0)
mnist_loader = get_mnist_dataloader(batch_size=BATCH_SIZE)
model = torch.nn.Sequential(
torch.nn.Conv2d(1, 20, 5, 1),
torch.nn.ReLU(),
torch.nn.MaxPool2d(2, 2),
torch.nn.Conv2d(20, 50, 5, 1),
torch.nn.ReLU(),
torch.nn.MaxPool2d(2, 2),
torch.nn.Flatten(),
torch.nn.Linear(4 * 4 * 50, 500),
torch.nn.ReLU(),
torch.nn.Linear(500, 10),
).to(DEVICE)
loss_function = torch.nn.CrossEntropyLoss().to(DEVICE)
def get_accuracy(output, targets):
"""Helper function to print the accuracy"""
predictions = output.argmax(dim=1, keepdim=True).view_as(targets)
return predictions.eq(targets).float().mean().item()
Writing the optimizer
To compute the update, we will need access to the diagonal of the Gauss-Newton,
which will be provided by Backpack in the diag_ggn_mc field,
in addition to the grad field created py PyTorch.
We can use it to compute the update direction
for a parameter p as
class DiagGGNOptimizer(torch.optim.Optimizer):
def __init__(self, parameters, step_size, damping):
super().__init__(parameters, dict(step_size=step_size, damping=damping))
def step(self):
for group in self.param_groups:
for p in group["params"]:
step_direction = p.grad / (p.diag_ggn_mc + group["damping"])
p.data.add_(step_direction, alpha=-group["step_size"])
Running and plotting
After extend-ing the model and the loss function and creating the optimizer,
the only difference with a standard PyTorch training loop will be the activation
of the DiagGGNMC` extension using a with backpack(DiagGGNMC()): block,
so that BackPACK stores the diagonal of the GGN in the
diag_ggn_mc field during the backward pass.
extend(model)
extend(loss_function)
optimizer = DiagGGNOptimizer(model.parameters(), step_size=STEP_SIZE, damping=DAMPING)
losses = []
accuracies = []
for batch_idx, (x, y) in enumerate(mnist_loader):
optimizer.zero_grad()
x, y = x.to(DEVICE), y.to(DEVICE)
model.zero_grad()
outputs = model(x)
loss = loss_function(outputs, y)
with backpack(DiagGGNMC()):
loss.backward()
optimizer.step()
# Logging
losses.append(loss.detach().item())
accuracies.append(get_accuracy(outputs, y))
if (batch_idx % PRINT_EVERY) == 0:
print(
"Iteration %3.d/%3.d " % (batch_idx, MAX_ITER)
+ "Minibatch Loss %.3f " % losses[-1]
+ "Accuracy %.3f" % accuracies[-1]
)
if MAX_ITER is not None and batch_idx > MAX_ITER:
break
fig = plt.figure()
axes = [fig.add_subplot(1, 2, 1), fig.add_subplot(1, 2, 2)]
axes[0].plot(losses)
axes[0].set_title("Loss")
axes[0].set_xlabel("Iteration")
axes[1].plot(accuracies)
axes[1].set_title("Accuracy")
axes[1].set_xlabel("Iteration")
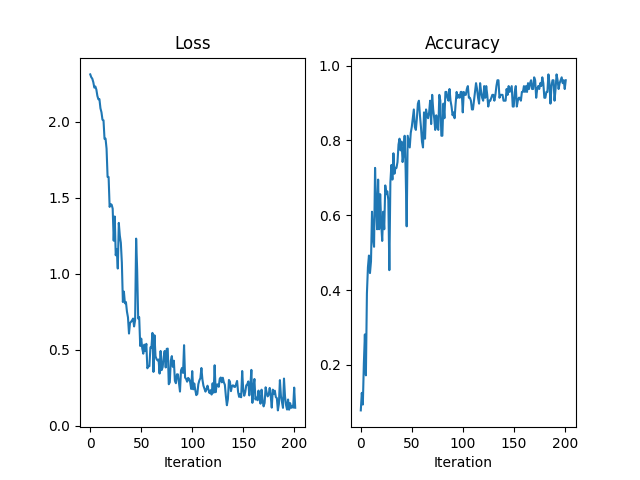
Iteration 0/200 Minibatch Loss 2.313 Accuracy 0.078
Iteration 50/200 Minibatch Loss 0.574 Accuracy 0.836
Iteration 100/200 Minibatch Loss 0.360 Accuracy 0.875
Iteration 150/200 Minibatch Loss 0.240 Accuracy 0.891
Iteration 200/200 Minibatch Loss 0.252 Accuracy 0.938
Text(0.5, 23.52222222222222, 'Iteration')
Total running time of the script: ( 0 minutes 28.623 seconds)